Resampling methods approximate the sampling distribution of a statistic or estimator. In essence, a sample taken from the population is treated as a population itself. A large number of new samples, or resamples, are taken from this “new population”, commonly with replacement, and within each of these resamples, the estimate of interest is re-obtained. A large number of these estimate replicates can then be used to construct the empirical sampling distribution from which confidence intervals, bias, and variance may be estimated. These methods are particularly advantageous for statistics or estimators for which no standard methods apply or are difficult to derive.
The jackknife is a popular resampling method, first introduced by Quenouille in 1949 as a method of bias estimation. In 1958, jackknifing was both named by Tukey and expanded to include variance estimation. A jackknife is a multipurpose tool, similar to a swiss army knife, that can get its user out of tricky situations. Efron later developed the arguably most popular resampling method, the bootstrap, in 1979 after being inspired by the jackknife.
In Efron’s (1982) book The jackknife, the bootstrap, and other resampling plans, he states,
Good simple ideas, of which the jackknife is a prime example, are our most precious intellectual commodity, so there is no need to apologize for the easy mathematical level.
Despite existing since the 1940’s, resampling methods were infeasible due to the computational power required to perform resampling and recalculate estimates many times. With today’s computing power, the uncomplicated yet powerful jackknife, and resampling methods more generally, should be a tool in every analyst’s toolbox.
Leave-one-out jackknife algorithm
The use of different resampling schemes results in different resampling methods. For example, bootstrap methods commonly sample ![]() observations with replacement from the original sample of size
observations with replacement from the original sample of size ![]() , repeating this a large number of
, repeating this a large number of ![]() times to obtain a large number of replicates.
times to obtain a large number of replicates.
The leave-one-out (LOO) jackknife creates ![]() samples of size
samples of size ![]() from the original sample of size
from the original sample of size ![]() by deleting each observation in turn. The leave-one-out jackknife is similar to the concept of leave-one-out crossvalidation encountered in prediction modelling or machine learning. More than one observation may be deleted at a time, though less common, in which case the method may be referred to as the leave-K-out jackknife.
by deleting each observation in turn. The leave-one-out jackknife is similar to the concept of leave-one-out crossvalidation encountered in prediction modelling or machine learning. More than one observation may be deleted at a time, though less common, in which case the method may be referred to as the leave-K-out jackknife.
The leave-one-out jackknife algorithm proceeds generally as follows:
- Estimate the parameter or estimand of interest
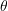 using the original sample of size
using the original sample of size  , referring to the resulting estimate as the full-sample estimate and denoting it by
, referring to the resulting estimate as the full-sample estimate and denoting it by 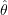 .
. - Construct
 jackknife samples of size
jackknife samples of size  by deleting each observation in turn. That is,
by deleting each observation in turn. That is,
(1)
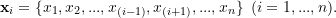
- Estimate the parameter or estimand of interest within each jackknife sample, yielding
 jackknife replicates,
jackknife replicates,
(2)
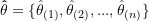
where subscript
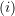 is used to indicate exclusion of the
is used to indicate exclusion of the 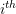 sample observation.
sample observation. - Obtain the jackknife estimate
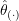 as the mean of the jackknife replicates,
as the mean of the jackknife replicates,
(3)
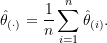
Bias estimation and adjustment
As noted, the jackknife was introduced by Quenouille in 1949 as a method of estimating bias.
Suppose ![]() is the full-sample estimate based on the entire sample of size
is the full-sample estimate based on the entire sample of size ![]() and
and ![]() is our jackknife estimate obtained according to the algorithm in the previous section. The jackknife estimate of the bias, also known as Quenouille’s estimate, is then obtained as
is our jackknife estimate obtained according to the algorithm in the previous section. The jackknife estimate of the bias, also known as Quenouille’s estimate, is then obtained as
(4) ![]()
A bias-corrected jackknife estimate is then obtained as,
(5) ![]()
If the full-sample estimate is unbiased such that ![]() , it follows that
, it follows that ![]() and the bias-corrected jackknife estimate
and the bias-corrected jackknife estimate ![]() is equivalent to the full-sample estimate
is equivalent to the full-sample estimate ![]() and jackknifing provides no advantages with respect to bias.
and jackknifing provides no advantages with respect to bias.
If the full-sample estimate is biased such that ![]() , the bias-corrected jackknife estimate will always reduce the bias compared to the full-sample estimate! However, the bias will not always be reduced to zero depending on its structure.
, the bias-corrected jackknife estimate will always reduce the bias compared to the full-sample estimate! However, the bias will not always be reduced to zero depending on its structure.
Suppose we assume that for a sample of size ![]() ,
, ![]() , the full-sample estimate
, the full-sample estimate ![]() is biased such that its expectation takes the form,
is biased such that its expectation takes the form,
(6) ![]()
where the ![]() do not depend on the sample size
do not depend on the sample size ![]() . Then, since each jackknife replicate is constructed using a jackknife sample of size
. Then, since each jackknife replicate is constructed using a jackknife sample of size ![]() , it follows that the expectation of the jackknife estimate is given by,
, it follows that the expectation of the jackknife estimate is given by,
(7) ![Rendered by QuickLaTeX.com \begin{align*} \mathbb{E}_{F}\left[ \hat{\theta}_{(\cdot)} \right] &= \frac{1}{n} \sum_{i=1}^{n} \mathbb{E}_{F} \left[ \hat{\theta}_{(i)} \right] \\ &= \frac{1}{n} \sum_{i=1}^{n} \left[ \theta + \frac{a_1(F)}{n-1} + \frac{a_2(F)}{(n-1)^2} + ... \right] \\ &= E_{n-1}. \end{align*}](https://statisticelle.com/wp-content/ql-cache/quicklatex.com-d02e2ce923cb65ab5c05153e04b84b81_l3.png)
That is, the expectation of the jackknife estimate is equivalent to the expectation of estimator of interest based on a sample of size ![]() . The expected difference between the full-sample estimate and the jackknife estimate is then,
. The expected difference between the full-sample estimate and the jackknife estimate is then,
(8) ![]()
If the assumed bias structure is correct, ![]() approximates the first bias term of
approximates the first bias term of ![]() . However, if the bias structure features higher order bias terms, extensions of the jackknife, such as Quenouille’s second-order jackknife, may be required.
. However, if the bias structure features higher order bias terms, extensions of the jackknife, such as Quenouille’s second-order jackknife, may be required.
Pseudo-observations and variance estimation
Tukey (1958) noticed that the bias-corrected jackknife estimate could be expressed as the mean of ![]() pseudo-observations, defined for the
pseudo-observations, defined for the ![]() sample observation as,
sample observation as,
(9) ![]()
such that,
(10) ![]()
The ![]() pseudo-observation can be viewed or interpretted as the
pseudo-observation can be viewed or interpretted as the ![]() individual’s contribution to the estimate.
individual’s contribution to the estimate.
Tukey further conjectured that pseudo-observations could be treated as independent and identically distributed random variables. When true, estimating the variance of the bias-corrected jackknife estimate ![]() is equivalent to estimating the variance of mean of the pseudo-observations. That is,
is equivalent to estimating the variance of mean of the pseudo-observations. That is,
(11) ![]()
If ![]() is assumed to be asymptotically Normally distributed, confidence intervals for
is assumed to be asymptotically Normally distributed, confidence intervals for ![]() can be constructed in the usual manner as,
can be constructed in the usual manner as,
(12) ![]()
Tukey’s conjectures were published in a short abstract with no supporting proofs. Miller (1968, 1974) later demonstrated that the properties conjectured only held for particular classes of statistics. However, this class of statistics does appear to include expectation functionals or linear functionals (among others)!
For more detail on expectation functionals and their estimators, check out my blog post U-, V-, and Dupree statistics.
- Tukey, J. W. (1958). Bias and confidence in not-quite large samples (Abstract). The Annals of Mathematical Statistics, 29(2), 614. doi:10.1214/aoms/1177706647
- Miller, R. G. (1968). Jackknifing Variances. The Annals of Mathematical Statistics, 39(2), 567–582. doi:10.1214/aoms/1177698418
- Miller, R. G. (1974). The Jackknife–A Review. Biometrika, 61(1), 1-15. doi:10.2307/2334280]
Example: Biased variance estimator
Data generation and estimator properties
In the following section, we demonstrate properties of the discussed jackknife estimators and pseudo-observations for the biased estimator of the variance with denominator ![]() ,
,
(13) ![]()
The expectation of ![]() takes the form of
takes the form of ![]() described within the Bias section,
described within the Bias section,
(14) ![]()
with ![]() , and it follows that,
, and it follows that,
(15) ![]()
When the ![]() are independent and identically distributed according to
are independent and identically distributed according to ![]() ,
,
(16) ![]()
Here, we generate a sample of ![]() independent and identically distributed observations generated according to
independent and identically distributed observations generated according to ![]() .
.
set.seed(12345)
n <- 100
mu <- 100
sigma <- 7
x <- rnorm(n, mu, sigma)
For reference, the first five generated observations are: 104.1, 105, 99.2, 96.8, 104.2.
A histogram of all ![]() generated observations is provided below.
generated observations is provided below.
qplot(x, geom='histogram', bins=5) + theme_bw()
It follows that the true properties of ![]() in this scenario are as follows.
in this scenario are as follows.
trueBias <- -sigma^2/n
trueBias
## [1] -0.49
trueVar <- 2 * (n-1) * sigma^4 / n^2
trueVar
## [1] 47.5398
That is, ![]() = -0.49 and
= -0.49 and ![]() = 47.54.
= 47.54.
Leave-one-out jackknife algorithm
First, we obtain the full-sample estimate by applying a given Estimator to the sample x.
getFullEst <- function(x, Estimator){
Estimator(x)
}
In this example, we are using the biased variance estimator so that Estimator = bVar.
bVar <- function(x){
n <- length(x)
bVar<- 1/n * sum((x-mean(x))^2)
return(bVar)
}
FullEst <- getFullEst(x, bVar)
Thus, the full-sample estimate is ![]() 60.28.
60.28.
Second, we use a loop to construct a list of the ![]() jackknife
jackknife Samples of size ![]() by deleting each observation in turn.
by deleting each observation in turn.
getSamples <- function(x){
n <- length(x)
Samples <- list()
for(i in 1:n){
Samples[[i]] <- x[-i]
}
return(Samples)
}
Samples <- getSamples(x)
Inspecting the first five observations of the first sample, we can see that ![]() 104.1 has been deleted: 105, 99.2, 96.8, 104.2, 87.3, 104.4.
104.1 has been deleted: 105, 99.2, 96.8, 104.2, 87.3, 104.4.
Third, we use sapply to apply Estimator to each of the ![]()
Samples and store the ![]() resulting
resulting ![]() in a vector
in a vector Replicates.
getReplicates <- function(Samples, Estimator){
Replicates <- sapply(Samples, FUN=Estimator, simplify=T)
return(Replicates)
}
Replicates <- getReplicates(Samples, bVar)
The first ten replicates, ![]() , are: 60.83, 60.78, 60.83, 60.64, 60.82, 58.76, 60.81, 60.75, 60.75, 60.21.
, are: 60.83, 60.78, 60.83, 60.64, 60.82, 58.76, 60.81, 60.75, 60.75, 60.21.
Fourth, we take the average of all jackknife Replicates to obtain the jackknife estimate JackEst.
getJackEst <- function(Replicates){
n <- length(Replicates)
JackEst <- mean(Replicates)
return(list(JackEst=JackEst, n=n))
}
JackEst <- getJackEst(Replicates)
The resulting value of ![]() = 60.27.
= 60.27.
Bias estimation and adjustment
To obtain a bias-corrected jackknife estimate, ![]() , we first need to obtain an estimate of the bias.
, we first need to obtain an estimate of the bias.
getBias <- function(FullEst, JackEst){
n <- JackEst$n
JackEst <- JackEst$JackEst
Bias <- (n-1) * (JackEst - FullEst)
return(Bias)
}
Bias <- getBias(FullEst, JackEst)
The estimated bias of the full-sample estimate ![]() is -0.609. Recall that the true bias
is -0.609. Recall that the true bias ![]() is -0.49.
is -0.49.
getAdjEst <- function(FullEst, Bias){
AdjEst <- FullEst - Bias
return(AdjEst)
}
AdjEst <- getAdjEst(FullEst, Bias)
Subtracting the estimated bias from the full-sample estimate yields a bias-adjusted jackknife estimate ![]() 60.89. Recall that the true value of
60.89. Recall that the true value of ![]() and the full-sample estimate
and the full-sample estimate ![]() 60.28.
60.28.
Pseudo-observations and variance estimation
To estimate the variance of our bias-adjusted jackknife estimate ![]() , we first need to transform each observation
, we first need to transform each observation ![]() into a pseudo-observation
into a pseudo-observation ![]() .
.
getPseudoObs <- function(FullEst, Replicates){
n <- length(Replicates)
PseudoObs <- n*FullEst - (n-1)*Replicates
return(PseudoObs)
}
PseudoObs <- getPseudoObs(FullEst, Replicates)
The first five pseudoobservations ![]() are: 5.73, 10.67, 6.22, 24.16, 6.44.
are: 5.73, 10.67, 6.22, 24.16, 6.44.
Finally, we obtain an estimate of the variance of ![]() as the sample variance of the sample mean of the pseudo-observations.
as the sample variance of the sample mean of the pseudo-observations.
getVarAdj <- function(PseudoObs){
n <- length(PseudoObs)
Var <- (1/n) * var(PseudoObs)
return(Var)
}
VarAdj <- getVarAdj(PseudoObs)
The estimated variance of the bias-adjusted jackknife estimate ![]() is 53.89. Recall that the true value
is 53.89. Recall that the true value ![]() 47.54.
47.54.
Summary
Based on our single sample of ![]() i.i.d observations from
i.i.d observations from ![]() , the jackknife appears to have provided a reasonable estimate of the parameter of interest
, the jackknife appears to have provided a reasonable estimate of the parameter of interest ![]() (est. = 60.89), the bias of the estimator = -0.49 (est. -0.61), and the variance of the estimator = 47.54 (est. = 53.89).
(est. = 60.89), the bias of the estimator = -0.49 (est. -0.61), and the variance of the estimator = 47.54 (est. = 53.89).
Discrepancies between true and estimated values are likely due to sampling variation (and the use of a single sample to evaluate the methods). If we repeat this process for 1,000 samples of size ![]() from
from ![]() and average the results across the samples, we would expect close approximation of the true values.
and average the results across the samples, we would expect close approximation of the true values.
results <- data.frame(Bias=NA, AdjEst=NA, VarAdj=NA)
n_sim <- 1000
for(s in 1:n_sim){
x <- rnorm(n, mu, sigma)
FullEst <- getFullEst(x, bVar)
Samples <- getSamples(x)
Replicates <- getReplicates(Samples, bVar)
JackEst <- getJackEst(Replicates)
Bias <- getBias(FullEst, JackEst)
AdjEst <- getAdjEst(FullEst, Bias)
PseudoObs <- getPseudoObs(FullEst, Replicates)
VarAdj <- getVarAdj(PseudoObs)
results[s,] <- c(Bias, AdjEst, VarAdj)
}
round(colMeans(results), 2)
## Bias AdjEst VarAdj ## -0.49 49.01 48.32
Indeed, we recover the true value of the estimator, its bias, and variance almost exactly!

P.S. The mean (biased) full-sample estimate across the 1,000 samples was 48.52.
Hello! Very helpful post. Quick question to make sure I understand the utility of this approach in applied work where the true parameters are unknown. Is the utility of the jack-knife that I can apply this approach to any standard model (e.g., regression) in a single sample and report the average of the jack-knifed results, that mean should be a better estimate of the true, un-biased effects?
Hi Daniel,
Thanks for the great question. 😊
I think that the bias reduction property of jackknife estimators is interesting. However, I actually think the real value of the jackknife in modern applications is the construction of pseudo-observations and their use in variance estimation.
In the case of regression, I’m not so sure the jackknife with respect to bias would provide many advantages since most coefficient estimators are unbiased.
However, for estimands without regression frameworks, pseudo-observations can be treated as the response in a GLM to adjust for covariates and estimate treatment effects!
This paper provides some cool examples of the application of pseudo-observations in survival analysis:
Andersen, P. K., & Pohar Perme, M. (2009). Pseudo-observations in survival analysis. Statistical Methods in Medical Research, 19(1), 71–99. doi:10.1177/0962280209105020
I’m hoping to follow-up this blog post with another discussing some applications of pseudo-observations… eventually…
Cheers,
Emma
very fascinating, thank you!