After my last grand slam title, U-, V-, and Dupree statistics I was really feeling the pressure to keep my title game strong. Thank you to my wonderful friend Steve Lee for suggesting this beautiful title.
Overview
A statistical functional is any real-valued function of a distribution function ![]() such that
such that
![]()
and represents characteristics of the distribution ![]() and include the mean, variance, and quantiles.
and include the mean, variance, and quantiles.
Often times ![]() is unknown but is assumed to belong to a broad class of distribution functions
is unknown but is assumed to belong to a broad class of distribution functions ![]() subject only to mild restrictions such as continuity or existence of specific moments.
subject only to mild restrictions such as continuity or existence of specific moments.
A random sample ![]() can be used to construct the empirical cumulative distribution function (ECDF)
can be used to construct the empirical cumulative distribution function (ECDF) ![]() ,
,
![]()
which assigns mass ![]() to each
to each ![]() .
.
![]() is a valid, discrete CDF which can be substituted for
is a valid, discrete CDF which can be substituted for ![]() to obtain
to obtain ![]() . These estimators are referred to as plug-in estimators for obvious reasons.
. These estimators are referred to as plug-in estimators for obvious reasons.
For more details on statistical functionals and plug-in estimators, you can check out my blog post Plug-in estimators of statistical functionals!
Many statistical functionals take the form of an expectation of a real-valued function ![]() with respect to
with respect to ![]() such that for
such that for ![]() ,
,
![]()
When ![]() is a function symmetric in its arguments such that, for e.g.
is a function symmetric in its arguments such that, for e.g. ![]() , it is referred to as a symmetric kernel of degree
, it is referred to as a symmetric kernel of degree ![]() . If
. If ![]() is not symmetric, a symmetric equivalent
is not symmetric, a symmetric equivalent ![]() can always be found,
can always be found,
![]()
where ![]() represents the set of all permutations of the indices
represents the set of all permutations of the indices ![]() .
.
A statistical functional ![]() belongs to a special family of expectation functionals when:
belongs to a special family of expectation functionals when:
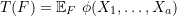 , and
, and is a symmetric kernel of degree
is a symmetric kernel of degree  .
.
Plug-in estimators of expectation functionals are referred to as V-statistics and can be expressed explicitly as,
![]()
so that ![]() is the average of
is the average of ![]() evaluated at all possible permutations of size
evaluated at all possible permutations of size ![]() from
from ![]() . Since the
. Since the ![]() can appear more than once within each summand,
can appear more than once within each summand, ![]() is generally biased.
is generally biased.
By restricting the summands to distinct indices only an unbiased estimator known as a U-statistic arises. In fact, when the family of distributions ![]() is large enough, it can be shown that a U-statistic can always be constructed for expectation functionals.
is large enough, it can be shown that a U-statistic can always be constructed for expectation functionals.
Since ![]() is symmetric, we can require that
is symmetric, we can require that ![]() , resulting in
, resulting in ![]() combinations of the subscripts
combinations of the subscripts ![]() . The U-statistic is then the average of
. The U-statistic is then the average of ![]() evaluated at all
evaluated at all ![]() distinct combinations of
distinct combinations of ![]() ,
,
![]()
While ![]() within each summand now, each
within each summand now, each ![]() still appears in multiple summands, suggesting that
still appears in multiple summands, suggesting that ![]() is the sum of correlated terms. As a result, the central limit theorem cannot be relied upon to determine the limiting distribution of
is the sum of correlated terms. As a result, the central limit theorem cannot be relied upon to determine the limiting distribution of ![]() .
.
For more details on expectation functionals and their estimators, you can check out my blog post U-, V-, and Dupree statistics!
This blog post provides a walk-through derivation of the limiting, or asymptotic, distribution of a single U-statistic ![]() .
.
Continue reading Getting to know U: the asymptotic distribution of a single U-statistic

