Parametric statistics assume that the unknown CDF ![]() belongs to a family of CDFs characterized by a parameter (vector)
belongs to a family of CDFs characterized by a parameter (vector) ![]() . As the form of
. As the form of ![]() is assumed, the target of estimation is its parameters
is assumed, the target of estimation is its parameters ![]() . Thus, all uncertainty about
. Thus, all uncertainty about ![]() is comprised of uncertainty about its parameters. Parameters are estimated by
is comprised of uncertainty about its parameters. Parameters are estimated by ![]() , and estimates are be substituted into the assumed distribution to conduct inference for the quantities of interest. If the assumed distribution
, and estimates are be substituted into the assumed distribution to conduct inference for the quantities of interest. If the assumed distribution ![]() is incorrect, inference may also be inaccurate, or trends in the data may be missed.
is incorrect, inference may also be inaccurate, or trends in the data may be missed.
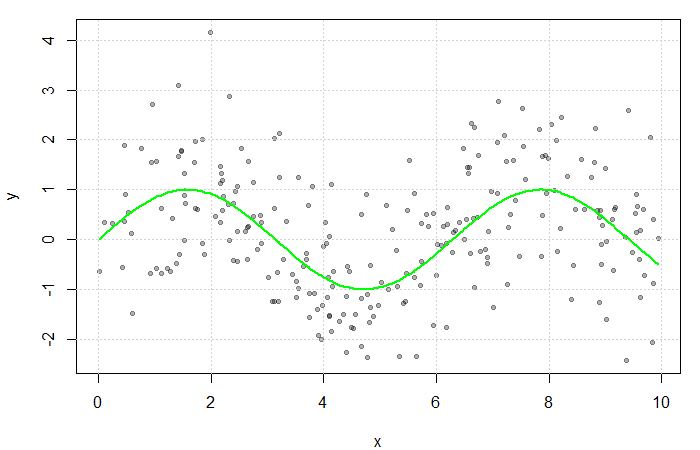
To demonstrate the parametric approach, consider ![]() independent and identically distributed random variables
independent and identically distributed random variables ![]() generated from an exponential distribution with rate
generated from an exponential distribution with rate ![]() . Investigators wish to estimate the 75
. Investigators wish to estimate the 75![]() percentile and erroneously assume that their data is normally distributed. Thus,
percentile and erroneously assume that their data is normally distributed. Thus, ![]() is assumed to be the Normal CDF but
is assumed to be the Normal CDF but ![]() and
and ![]() are unknown. The parameters
are unknown. The parameters ![]() and
and ![]() are estimated in their typical way by
are estimated in their typical way by ![]() and
and ![]() , respectively. Since the normal distribution belongs to the location-scale family, an estimate of the
, respectively. Since the normal distribution belongs to the location-scale family, an estimate of the ![]() percentile is provided by,
percentile is provided by,
![]()
where ![]() is the standard normal quantile function, also known as the probit.
is the standard normal quantile function, also known as the probit.
set.seed(12345)
library(tidyverse, quietly = T)
# Generate data from Exp(2)
x <- rexp(n = 100, rate = 2)
# True value of 75th percentile with rate = 2
true <- qexp(p = 0.75, rate = 2)
true
## [1] 0.6931472
# Estimate mu and sigma
xbar <- mean(x)
s <- sd(x)
# Estimate 75th percentile assuming mu = xbar and sigma = s
param_est <- xbar + s * qnorm(p = 0.75)
param_est
## [1] 0.8792925
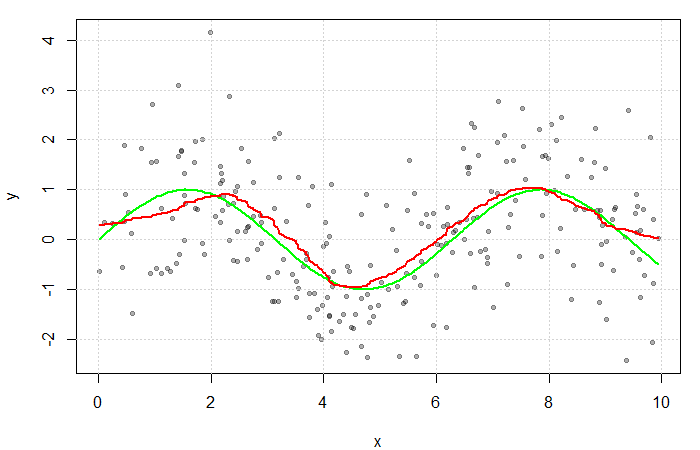
The true value of the 75![]() percentile of
percentile of ![]() is 0.69 while the parametric estimate is 0.88.
is 0.69 while the parametric estimate is 0.88.
Nonparametric statistics make fewer distributions about the unknown distribution ![]() , requiring only mild assumptions such as continuity or the existence of specific moments. Instead of estimating parameters of
, requiring only mild assumptions such as continuity or the existence of specific moments. Instead of estimating parameters of ![]() ,
, ![]() itself is the target of estimation.
itself is the target of estimation. ![]() is commonly estimated by the empirical cumulative distribution function (ECDF)
is commonly estimated by the empirical cumulative distribution function (ECDF) ![]() ,
,
![]()
Any statistic that can be expressed as a function of the CDF, known as a statistical functional and denoted ![]() , can be estimated by substituting
, can be estimated by substituting ![]() for
for ![]() . That is, plug-in estimators can be obtained as
. That is, plug-in estimators can be obtained as ![]() .
.
Continue reading Parametric vs. Nonparametric Approach to Estimations